Improving Photosynthesis
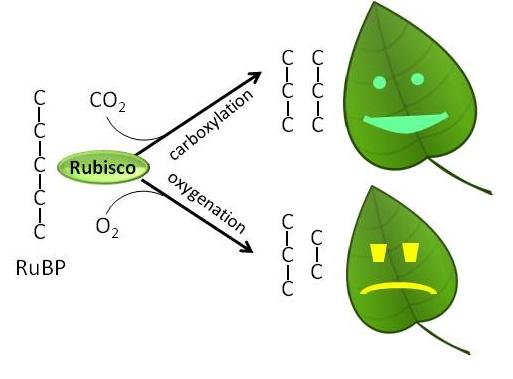
Yields of many crops could be increased if photosynthesis could be engineered to become more efficient. Our lab’s focus is on the plant carbon-fixing enzyme Rubisco (ribulose bisphosphate carboxylase/oxygenase). Supplying more CO2 to Rubisco has potential to improve carbon fixation. Furthermore, Rubisco's poor kinetic properties could potentially be improved through genetic engineering.
Project: Introducing a Cyanobacterial Carbon-concentrating Mechanism into Chloroplasts
One possible way to enhance carbon fixation by Rubisco is to surround the enzyme with CO2, a strategy that has evolved in cyanobacteria. These microorganisms contain microcompartments named carboxysomes that encapsulate Rubisco in a protein shell and contain an enzyme, carbonic anhydrase (CA), which converts bicarbonate to CO2. A typical microcompartment is 100-200 nm in size.
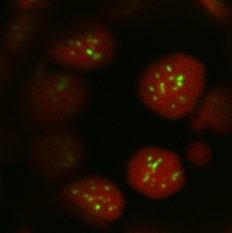
Image: Punctate loci obtained by expression and chloroplast targeting of multiple carboxysomal proteins
Red = chlorophyll autofluorescence
Green = YFP label on a carboxysomal protein
Cyanobacteria such as Synchococcus elongatus encode a Rubisco enzyme that is faster than land plant Rubisco, though more oxygen sensitive than plant enzymes. Because CO2 is concentrated near Rubisco within the carboxysome, the enzyme can work efficiently to fix carbon. We have been able to express β-cyanobacterial carboxysome shell proteins transiently in chloroplasts of tobacco leaves. By labeling one of the shell proteins with Yellow Fluorescent Protein (YFP), we were able to use confocal fluorescence microscopy to observe punctuate loci forming inside chloroplasts. Electron microscopy revealed the presence of both linear structures and oval structures similar to empty microcompartments.
We replaced the tobacco RbcL gene with transgenic loci containing two or three cyanobacterial genes encoding the S. elongatus Rubisco large subunit (Se LS), small subunit (Se SS) either alone or with the putative chaperone RbcX or a gene encoding the carboxysomal protein M35. Tobacco plants could grow phototrophically despite the absence of tobacco Rubisco, if provided with elevated CO2.
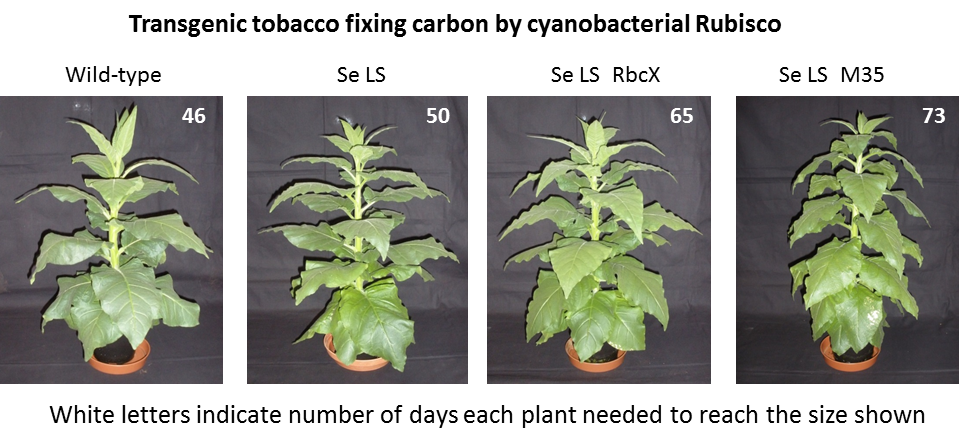
This unexpected finding was reported in a number of lay publications (as shown in our News section). In elevated CO2, the Se LS plants could grow nearly as fast as wild-type.
We are currently using chloroplast transformation to introduce the proteins needed to produce a functional carboxysome in plants. We are developing improved transformation technology to facilitate synthesis of multi-gene operons.
We are also engineering another component of the cyanobacterial carbon-concentrating mechanism. We have used CRISPR/Cas9 mutagenesis to remove chloroplast carbonic anhydrase activity. The next step is to engineer bicarbonate transporters into chloroplasts to supply substrate for use by Rubisco in carboxysomes. This project is led by postdoctoral associate Vishal Chaudhari.
We are collaborating with Martin Parry and Elizabete Carmo-Silva’s groups at Lancaster University, United Kingdom.
Work on this project is supported by Bilateral NSF/BIO-BBSRC:
Synthesis of Microcompartments in Plants for Enhanced Carbon Fixation, NSF Award 1642386 and NSF MCB 2131582 Systems and Synthetic Biology.
Project: Engineering Rubisco
The enzyme Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase) has been subjected to intensive analysis due to its key role in photosynthesis in converting atmospheric CO2 into energy-rich compounds. Rubisco exhibits a slow catalytic rate and can also react with O2, leading to photorespiration. In general, improvement in rate of catalysis results in loss of specificity, and vice versa. Nevertheless, there is variation between the properties of Rubisco in different species and phylogenetic groups. Mutational analysis has also revealed residues that can be manipulated and may improve one or the other features of the enzyme. During evolutionary time, the C3 plant enzyme has undergone changes to increase turnover rate or increase specificity for the reaction with CO2 instead of O2.
We have implemented an E. coli expression system that allows assembly of the Rubisco enzyme from the C3 plant Nicotiana tabacum (tobacco). We have improved the expression system for convenient production of mutant Rubisco, and have shown that native tobacco Rubisco sequences expressed in E. coli (eRubisco) produce an enzyme with a catalytic activity comparable to tobacco mesophyll Rubisco. We are producing mutated Rubisco in E. coli and are examining the effects of altered amino acids on enzyme kinetics and specificity. We intend to introduce mutated enzymes with interesting properties into plants through chloroplast transformation and examine the properties of photosynthesis in the transgenic plants.

Image: Trangenic tobacco plants containing mutant Rubisco.
This project is led by Myat Lin, postdoctoral Research Associate.
Our current work is supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences DE-SC0020142.




