RNA Editing in Plant Organelles
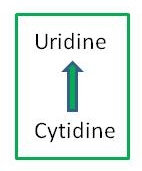
Particular cytidines in transcripts in chloroplasts and mitochondria of plant cells undergo modification to uridine by RNA editing. These C-to-U changes can create start and stop codons or change the encoded amino acid from the one predicted from the genomic sequence. RNA editing is known to occur in a variety of organisms, including mammals, insects, plants and microorganisms. In humans and other animals, RNA editing creates diversity in certain classes of proteins encoded by the nuclear genome. Editing in plants appears to correct defective RNA post-transcriptionally.
- Expressing plant editing factors and their target RNAs in E. coli to identify the composition of the editosome and its specificity for the correct C target.
- Mutagenesis of members of the OZ and ORMM editing factor families to determine their function
- Isolation of tagged editosomes by affinity methods followed by proteomic analysis of the preparations to detect novel components of editosomes
Past work in the Hanson lab on plant RNA editing
Cis-elements required for chloroplast RNA editing
Initially we characterized the elements present on the chloroplast transcripts that signaled which C was to be targeted for editing. We introduced sequences into the chloroplast genome that were mutated around the editing target to determine the signals for selection of the proper C.
We have also used chloroplast extracts capable of editing exogenous RNA to probe the features of transcripts that are important in RNA editing. Regions surrounding chloroplast editing sites were mutated in vitro and RNA transcribed from mutated templates. These RNAs could be introduced into chloroplast extracts and the extent of conversion of C to U was assayed.
Site-specific editing factors
Research in a variety of labs has shown that pentatricopeptide repeat (PPR)-containing proteins mediate specific recognition of the C target of editing in both chloroplasts and mitochondria. We have identified PPR protein RNA editing recognition factors by comparative genomics or by exploiting natural variation in editing extent.

We have identified the C targets recognized by several PPR proteins, such as RARE1 and QED1.
Some members of the PPR protein family contain a DYW domain at the C-terminus. This domain has been hypothesized to provide the enzymatic activity needed to convert C to U because it carries motifs characteristic of cytidine deaminases. We performed site-specific mutagenesis of these motifs in two PPR proteins and demonstrated that editing activity was lost. This finding supports the idea that DYW domains carry cytidine deaminase activity.

Sometimes the DYW domain can be provided in trans by a protein that does not carry enough information to recognize the cis-element. We characterized one such protein, MEF8, which has a very short set of PPR motifs plus a DYW domain, and is needed for editing at 38 mitochondrial C targets.
Identification of three editing factor families
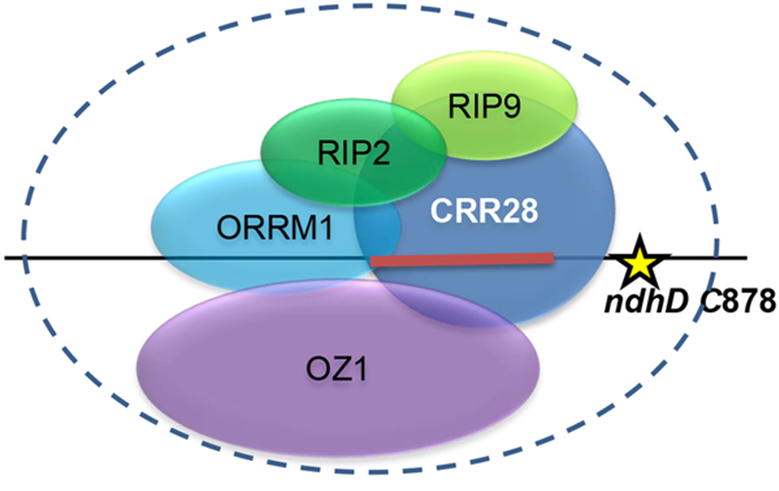
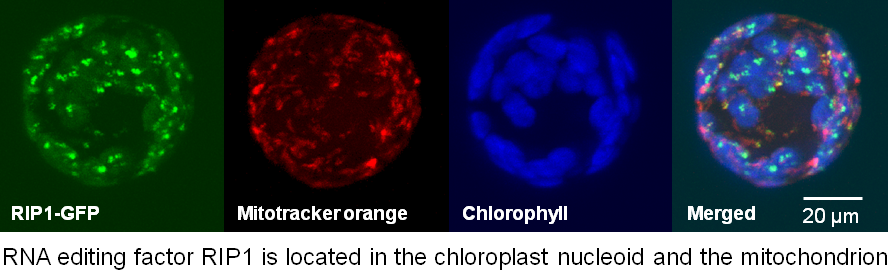
We identified the RIP family of editing factors by immunoprecipitation of a FLAG-tagged PPR protein. The RIP family (also called the MORF family), contains two factors needed for editing of many chloroplast C targets, two factors required for multiple mitochondrial targets, and one that is involved in both chloroplast and mitochondrial editing.
Homology search revealed that a protein that we have named ORRM1 (Organelle RRM-domain containing protein 1) contained domains characteristic of the RIP family, but also exhibited an RNA Recognition Motif (RRM), unlike any of the RIP proteins. An Arabidopsis orrm1 mutant exhibited defects in editing of multiple chloroplast editing sites. ORRM1 belongs to a clade within the large RRM family of Arabidopsis proteins.
By analyzing mutants and silenced tissue, we have subsequently demonstrated that other members of the ORRM clade are mitochondrial or chloroplast RNA editing factors.
We have identified another family required for RNA editing by immunoprecipitating ORRM1. We identified the protein Organelle Zinc Finger Protein 1 (OZ1) as required for editing of 14 targets in Arabidopsis.
Arabidopsis contains three additional members of the zinc finger family, all predicted to be targeted to organelles. The second member, OZ2, is not an editing factor—instead, a RNA splicing factor.
Further work should complete the picture of the multiple configurations of RNA editosomes which are required to convert Cs to Us in plant organelles. This knowledge has potential application in genetic engineering of plants and animals. By understanding how the natural process works, we may be able to engineer C-to-U changes into transcripts chosen for deliberate alteration.